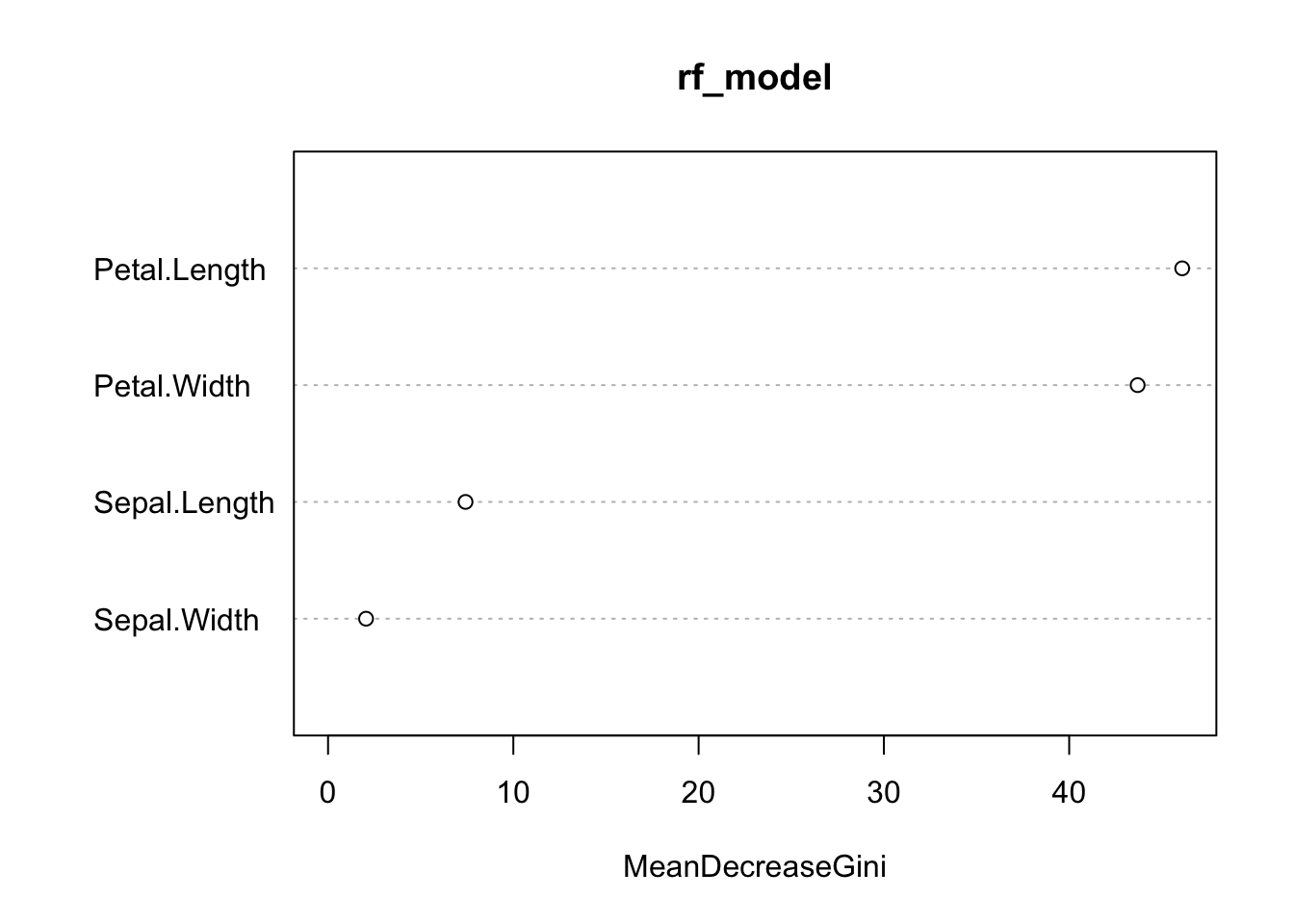
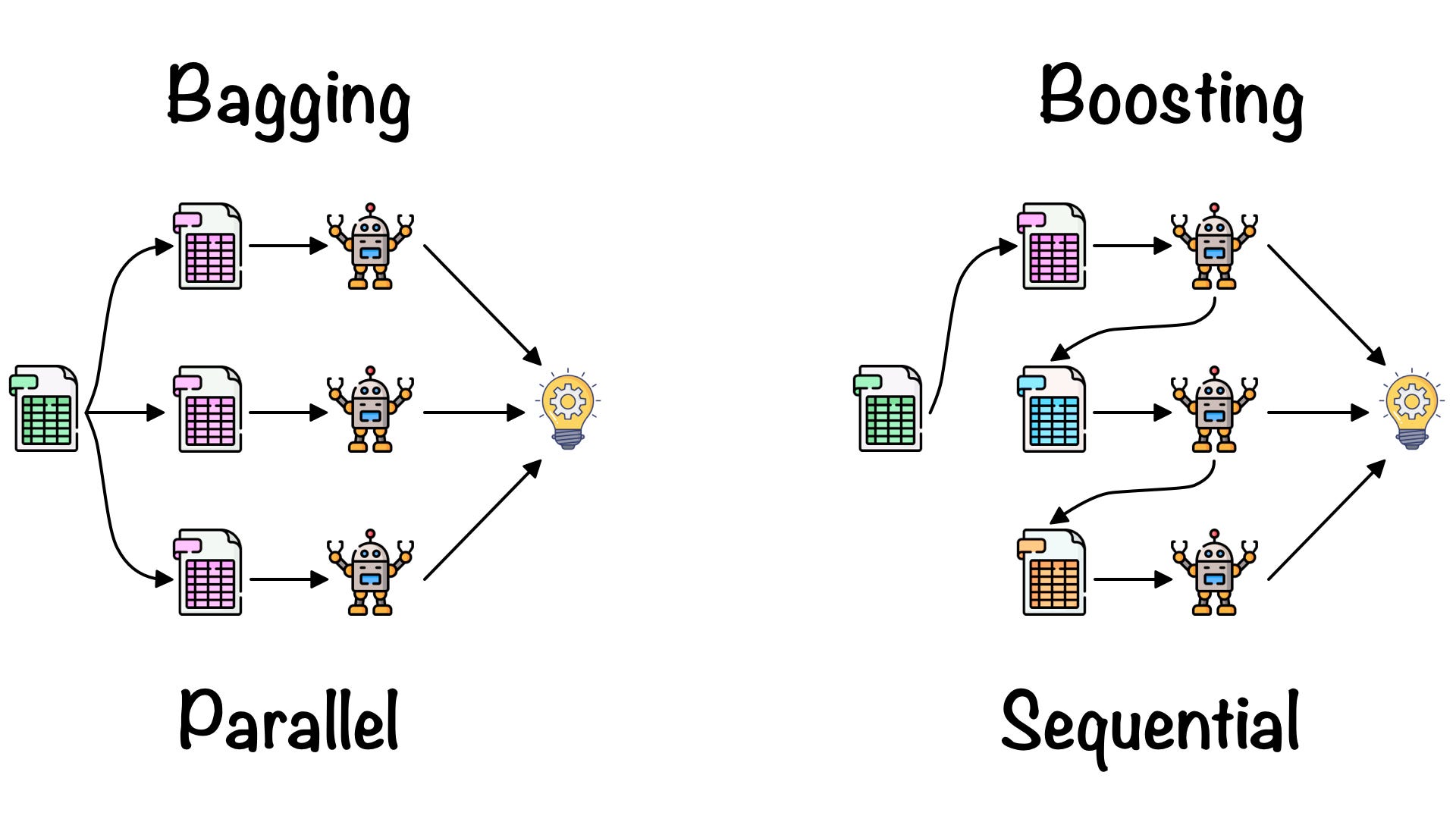
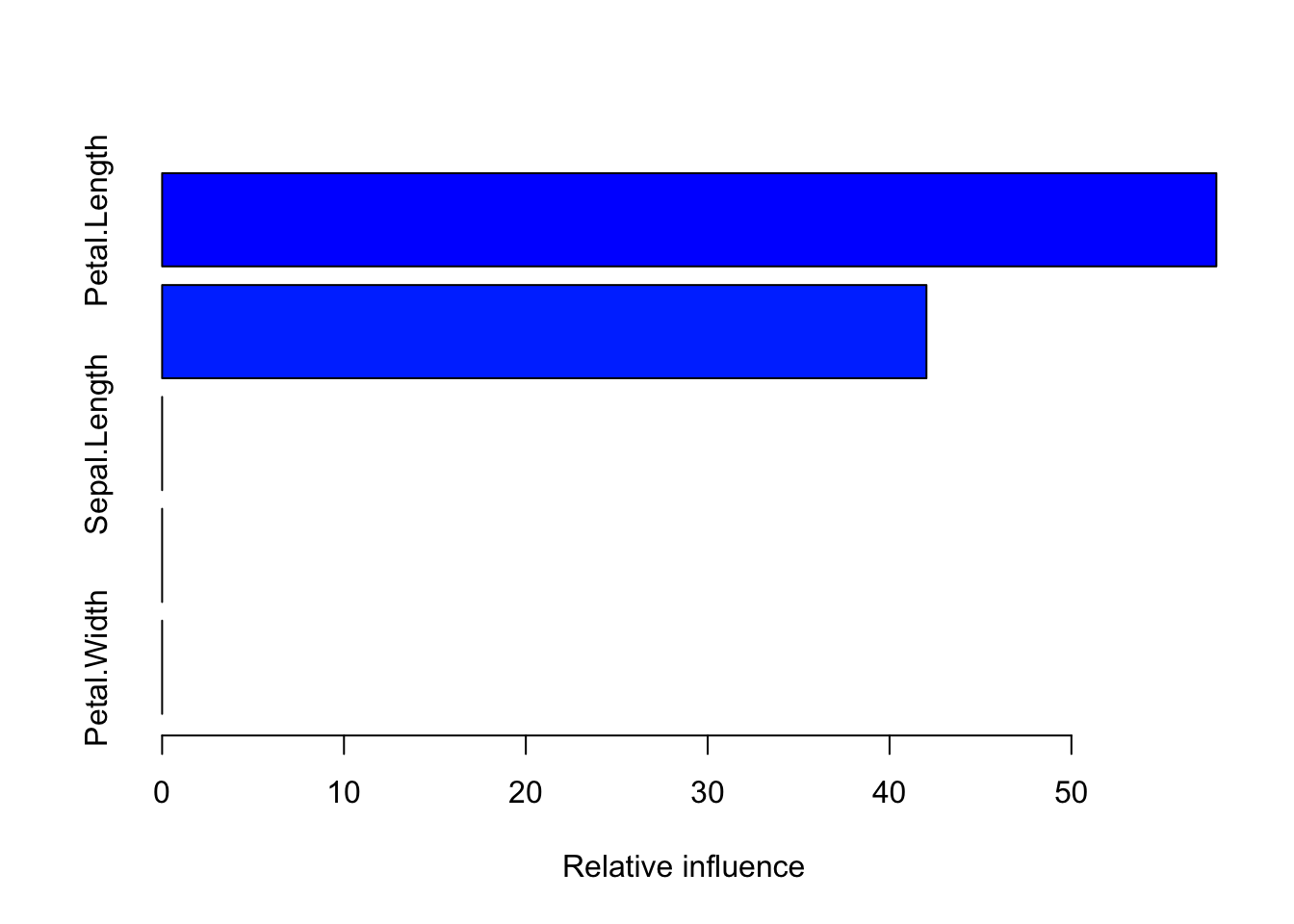
Imagine you’re playing a video game where you need to get past several levels, and each level is a bit harder than the last one. Now, suppose you’re not alone – you’ve got a group of friends helping you out. Each friend takes a turn to play a level. If they get through it, great! But if they don’t, the next friend pays extra attention to where things went wrong and tries to do better on that part.
In machine learning, “boosting” is like this team effort to win the game. Instead of friends, you have a group of helpers called “models” that try to predict something (like whether a photo shows an orange or an apple).
The first model makes its best guess, but it probably doesn’t get everything right. So, the next model watches and ‘learns’ from the mistakes of the first model, trying specifically to get those wrong predictions right, giving more focus to the harder parts. This process goes on with several models, each one learning from the errors of the one before it, and paying more attention to the things that are harder to predict.
In the end, all these models come together, combining their strengths, and make a final, super-smart guess. Just like how you and your friends would combine your gaming skills to win the video game, these models join forces to make a really good prediction.